Advanced Cardiovascular Imaging Lab, University of Pennsylvania
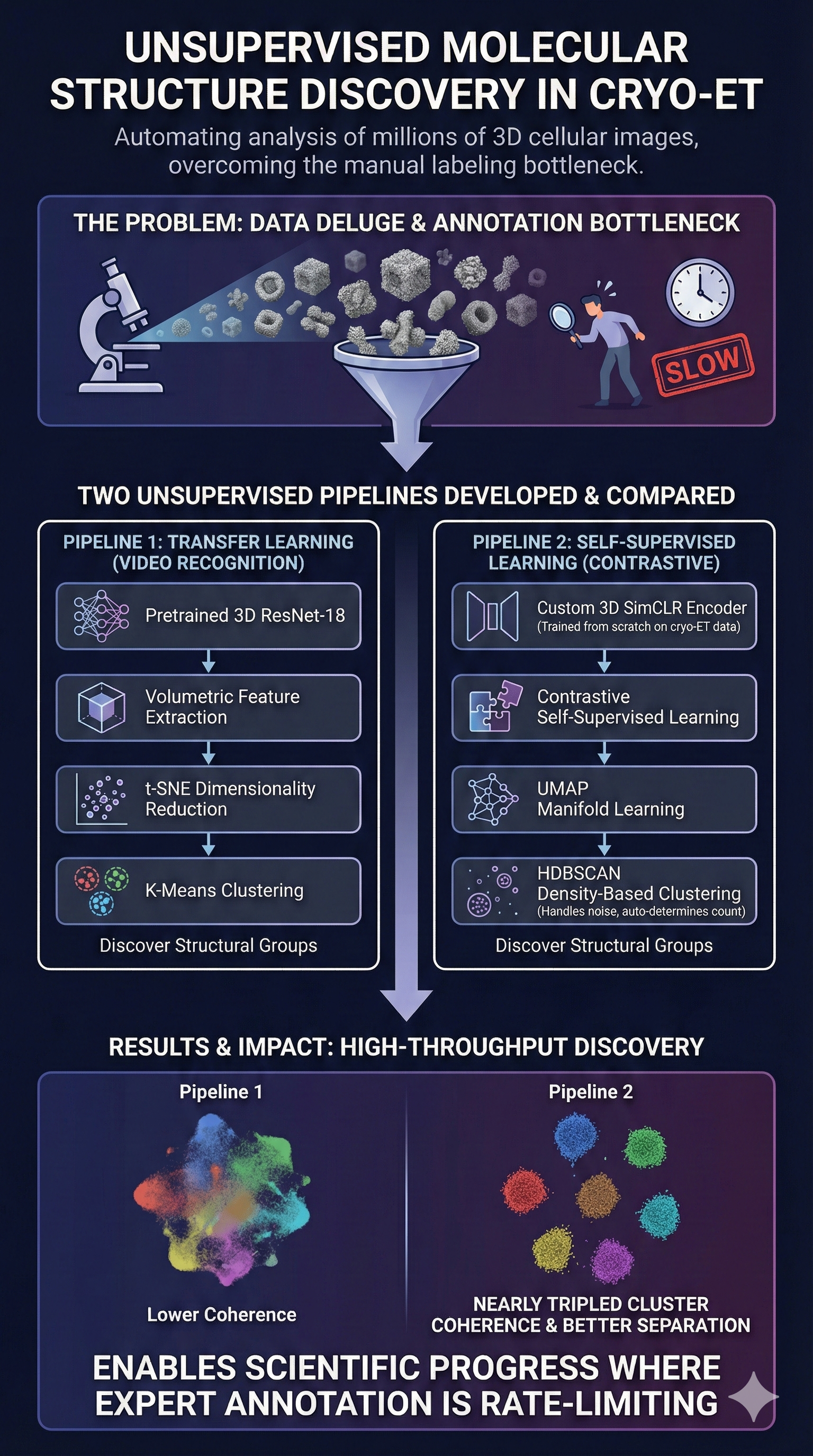
Hemothorax—internal chest bleeding from trauma—requires rapid diagnosis, but manual CT analysis is slow and subject to human variability. I developed an AI-assisted detection system at Penn Medicine to help radiologists identify these critical bleeds more quickly and consistently. The system combines multiple deep learning architectures working together: 2D models analyze slice-by-slice patterns while 3D models capture full volumetric context. By merging their predictions, this ensemble approach outperformed both individual models and fine-tuned versions of existing medical imaging systems. The work shows that architectural diversity, not just more data, can drive better clinical AI performance. This reduces diagnostic delays in emergency settings and gives radiologists a reliable AI assistant for detecting these life-threatening bleeds.